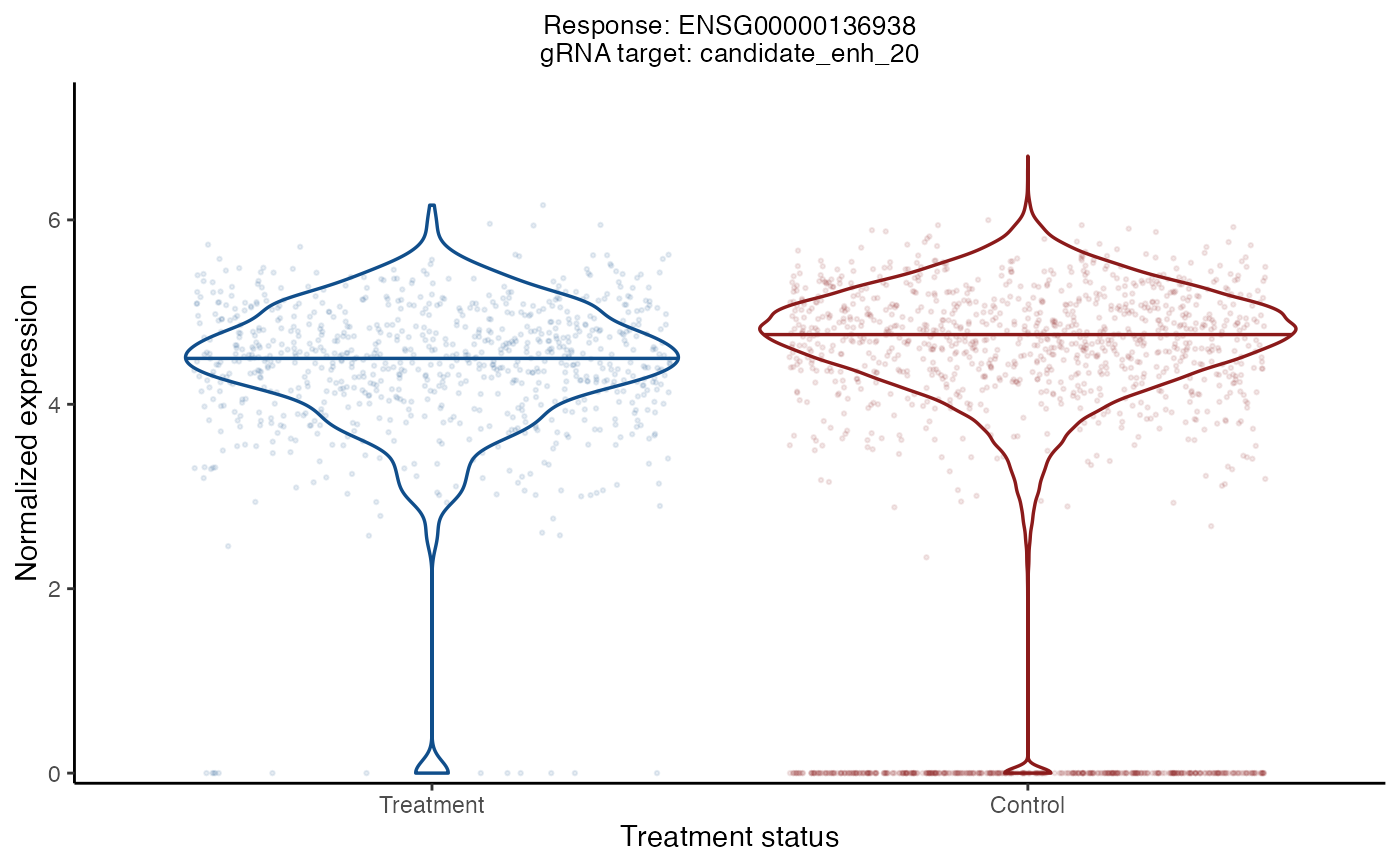
plot_response_grna_target_pair() creates a violin plot of the expression level of a given response as a function of the "treatment status" (i.e., treatment or control) of a given gRNA target. The left (resp., right) violin plot shows the expression level of the response in treatment (resp., control) cells. The expression level is normalized by dividing by n_response_umis, adding a pseudo-count of 1 and then taking the log transform. If the given response-gRNA-target pair has been analyzed, the p-value for the test of association also is displayed.
Arguments
- sceptre_object
a
sceptre_objectthat has hadrun_qc()called on it- response_id
a string containing a response ID
- grna_target
a string containing a gRNA target (or, if
grna_integration_strategyis set to"singleton", an individual gRNA ID)
Details
If grna_integration_strategy is set to "singleton", then grna_target should be set to a gRNA ID.
Examples
data(highmoi_example_data)
data(grna_target_data_frame_highmoi)
# import data
sceptre_object <- import_data(
response_matrix = highmoi_example_data$response_matrix,
grna_matrix = highmoi_example_data$grna_matrix,
grna_target_data_frame = grna_target_data_frame_highmoi,
moi = "high",
extra_covariates = highmoi_example_data$extra_covariates,
response_names = highmoi_example_data$gene_names
)
discovery_pairs <- construct_cis_pairs(sceptre_object)
sceptre_object |>
set_analysis_parameters(
side = "left",
discovery_pairs = discovery_pairs,
resampling_mechanism = "permutations",
) |>
assign_grnas(method = "thresholding") |>
run_qc() |>
run_discovery_analysis(
parallel = TRUE,
n_processors = 2
) |>
plot_response_grna_target_pair(
response_id = "ENSG00000211641",
grna_target = "candidate_enh_15"
)
#> Warning: The calibration check (`run_calibration_check()`) should be run before the discovery analysis.
#>
#> Generating permutation resamples. ✓
#> Running discovery_analysis in parallel. Change directories to /var/folders/wh/j0lkx_9d5zq75dljjxvvgb480000gn/T//RtmpzPqB2F/sceptre_logs/ and view the files discovery_analysis_*.out for progress updates.
#> ✓
#>